Coulomb Force predictions from chemical makeup
Python, Pandas, and Chemistry Project
Project Details
This is the final project for ECEN 489, an elective course in Computational Data Science. This class is taught by Dr. Jin Tao, and incredible researcher at the high computing center at Texas A&M.
Measurements at the atomic level are incredibly difficult, but of extreme relevance in the study of material sciences. One of the fundamental Measurements is that of the Coulomb Force. It is an equation that gives you a total force of 2 electrical charges affecting each other. It is straight forward for 2 atoms but increases in difficulty exponentially in 3D space and with multiple atoms in a molecule.
The question was then if we could create a prediction of total Coulomb force, based exclusively on the atomic formula of a molecule. This maybe wouldn’t be as accurate, but it may allow predictions for molecules so large and complex it is prohibitive to measure.
Process
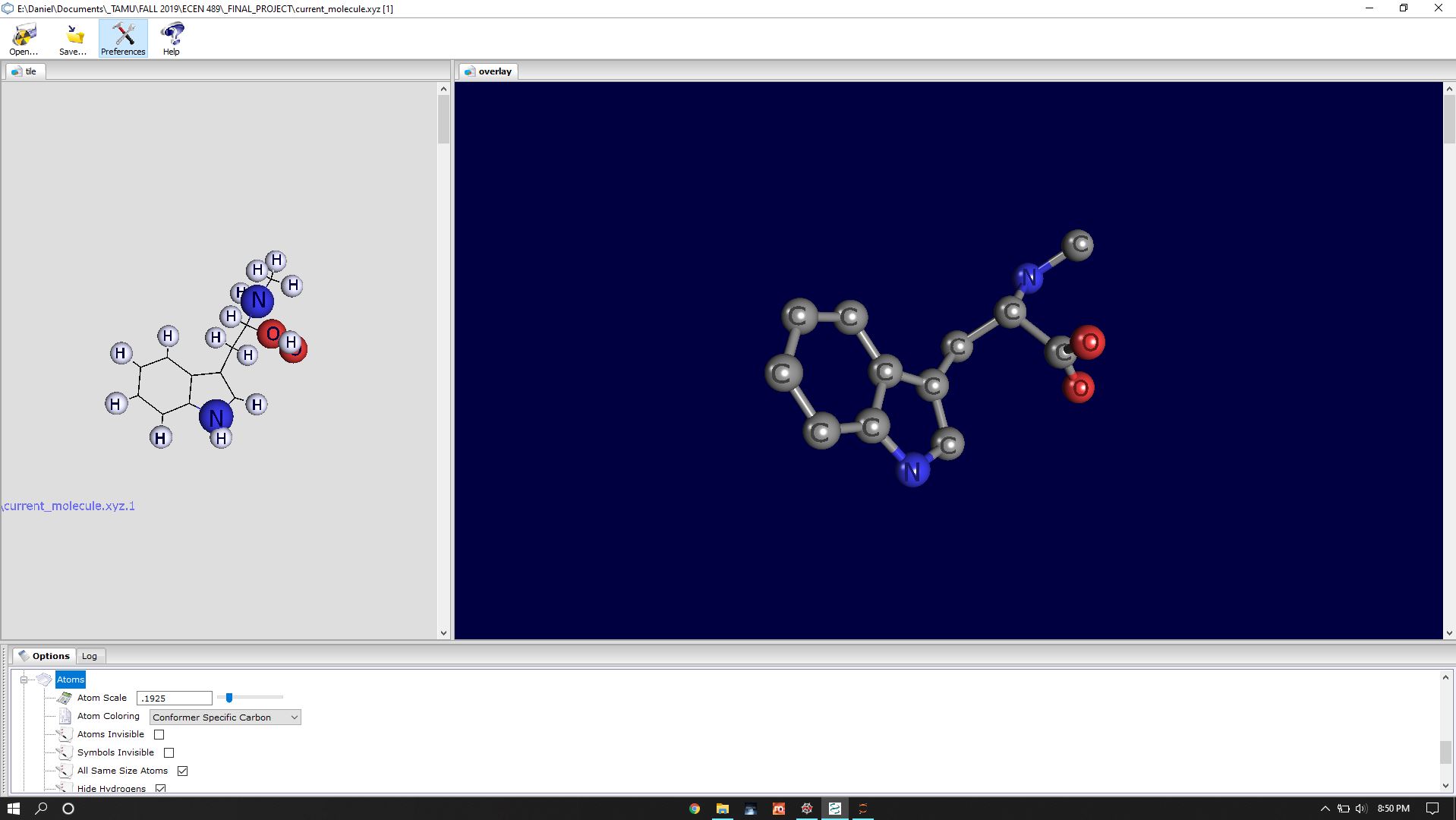
The first process is to visualize and understand the data. I used the PUBCHEM website to check my work and to obtain the data of simple molecules. The data was presented in JSON, and I used Pandas to import the data into DataFrames and extract what I needed. I then had to use a massive amount of string operators to create an XYZ file, which describes the chemical makeup of the molecule.
| Name | X | Y | Z | |
|---|---|---|---|---|
| O | -0.0782 | -1.5651000000000002 | 1.3894 | |
| O | -2.2297000000000002 | -0.9343 | -1.3664 | |
| O | 2.2983000000000002 | -1.2575 | -0.2803 | |
| O | 2.8296 | 0.9664 | -0.325 | |
| C | -0.30260000000000004 | -0.9556 | 0.1153 | |
| C | -1.8113000000000001 | -0.6388 | -0.0329 | |
| C | 0.5877 | 0.2525 | 0.013800000000000002 | |
| C | -2.18 | 0.7909 | 0.2585 | |
| C | 0.132 | 1.5099 | 0.1068 | |
| C | -1.268 | 1.77 | 0.33490000000000003 | |
| C | 2.0222 | 0.061700000000000005 | -0.2141 | |
| H | -0.041800000000000004 | -1.6945000000000001 | -0.6527000000000001 | |
| H | -2.3841 | -1.2908 | 0.6377 | |
| H | -3.2301 | 1.0276 | 0.4037 | |
| H | 0.7787000000000001 | 2.3781 | 0.0315 | |
| H | -1.5681 | 2.7917 | 0.54 | |
| H | 0.8454 | -1.8688 | 1.4156 | |
| H | -3.1904 | -0.7899 | -1.4159000000000002 | |
| H | 3.2524 | -1.4274 | -0.4325 |
Once I had the XYZ data, I used called PC3 to view individual molecules to check against the PubChem data.
With this Data, I can use an equation known as the Coulomb matrix equation to coulomb to create a matrix that represents the forces between in all of the atoms in a molecule. I then use the XYZ data I pulled to find if there was a comparison.
Conclusion
Ultimately, we ran out of time to complete a machine learning model, but this was a still pretty intense exercise in python and pandas. However, it is worth noting that there didn’t seem to be a direct connection from chemical makeup, to Coulomb force in a conventional correlation measurement. However, this was a project where I was able to pull data from a complex data set, clean it, and use operators to convert it into a file to be used by an open-source program.